_ Department of Nano Bioelectrical Laboratory (Nano Biosensors)
Electrochemical nano biosensors and a method called sandwich component Three assay
Researcher and author: Dr. ( Afshin Rashid)
Note: Electrochemical signal measurement methods are very suitable for detecting direct DNA oxidation because electrochemical reactions directly create an electronic signal and therefore there is no need for expensive converter devices.
In addition, in this process, because the sequence of the immobilized game can be limited to only a series of electrode substrates, the act of showing (tracking) is done by a series of inexpensive electrochemical analyzers. Electrochemical sensors are used to perform clinical or environmental tests; The sensitivity of electrochemical signals is based on the direct oxidation or catalysis of DNA spacers, as well as the reduction reactions of reporter molecules or enzymes. Various methods are used to measure the signal electrochemically. The basis of signal measurement in direct electrochemistry of DNA is based on the oxidation and reduction reaction of DNA in a mercury electrode, therefore, the amount of oxidized and reduced DNA is proportional to the amount of DNA that is hybridized with the probe. In addition to the old methods of direct DNA regeneration, a method called Stripping Adsorption Voltammetry is used for direct DNA oxidation, which is a very sensitive method. In the direct electrochemical method, purine is oxidized by materials such as: carbon, indium oxide (ITO), gold and polymer covered electrodes. Although the direct electrochemical method is a very sensitive method, its application is complicated because for the direct oxidation of DNA, a background current with a high potential is required. Also, advanced mathematical and numerical methods are necessary to be able to measure each signal. Of course, new methods have been designed by which the interference created in the field can be removed with the help of physical methods.
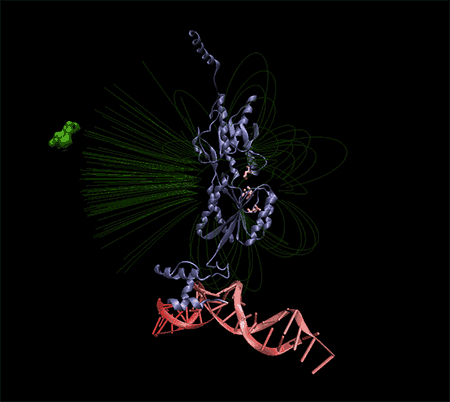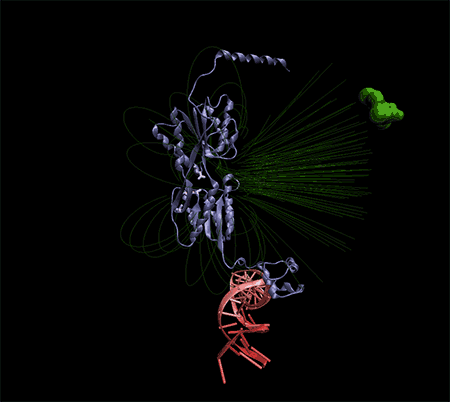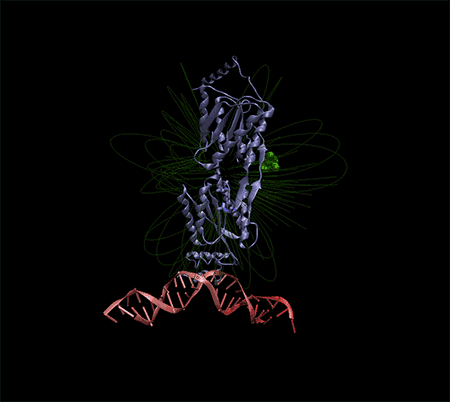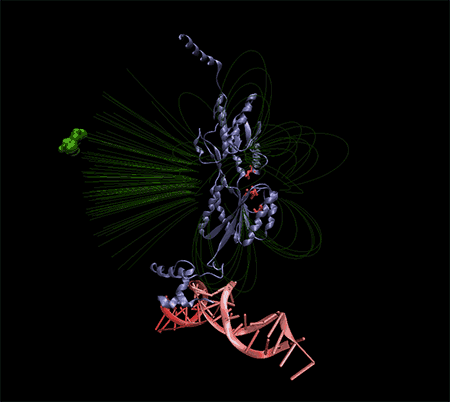
The target DNA is tracked by probes fixed on magnetic beads. In this method, after the probe DNA collides with the target DNA, the magnetic beads are separated in the solution. The collected DNAs are depurinated in an acidic solution, and free guanine and adenine nucleosides are collected and analyzed by absorption voltammetry. Also, a similar method using direct oxidation of guanine has been reported, by which they were able to identify a specific mutation among DNA fragments that were amplified by PCR. Another method is the use of peptide nucleic acid probes, with the use of these probes, hybridization is performed more firmly, and point mutations in the target DNA can be detected with this method.
_69nx.jpg)

In indirect DNA electrochemical method, electrochemical intermediates are used for DNA oxidation. A very effective specific method is the use of polypyridyl (RUII) and IIO complexes, which use their molecules to perform electrochemical oxidation of guanine. By this method, they trace a sequence of three nucleotides. This method together with PCR-rt can be used for the overexpression of genes in tumor samples and has high sensitivity. In the same method, the organic bases that have been chemically changed enter the PCR product and the resulting DNA fragments are identified. Although this method is very sensitive, it is still not clear whether this method is suitable for clinical diagnosis. One of the advantages of this method is that it is uncomplicated.
Conclusion :
There are several methods based on which the target DNA is tracked with molecule-labeled activated reporter molecules . By using physical methods, it is possible to determine the order of the labeled game up to the limit. This method includes the three-component sandwich assay (sandwich component Three assay), in which the regenerated labeled material is connected to a synthetic DNA fragment, which is this DNA fragment. It is connected to the probe-target complex . In other words, the reduced labeled material is connected to a synthetic fragment, which is attached or dangling to the probe-target complex. In this case, the need for a change in the target DNA is created, it is destroyed, which causes the probe and the labeled sequence to be placed together. One of the uses of labeled DNA is the use of a probe labeled with ferrosin, which is placed on a gold electrode. Using this technique, the target DNA can be enclosed, and due to the contact, a change is made in the ferrocene molecule, which gives a signal to the gold electrode, and this signal is measured as a Farad current.
Researcher and author: Dr. ( Afshin Rashid)
Specialized doctorate in nano-microelectronics