_ Department of Nano Bioelectrical Laboratory (Nano Biosensors)
All molecular-based electrochemical nano-biosensors depend on a very specific system to detect or track the target molecule
Researcher and author: Dr. ( Afshin Rashid)
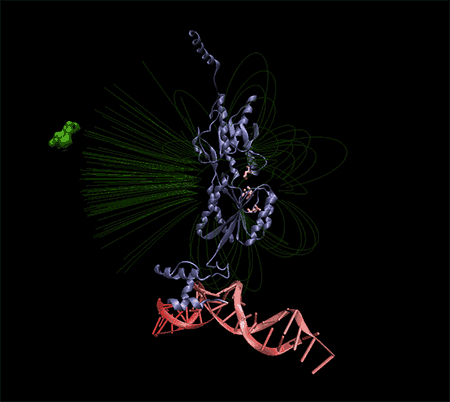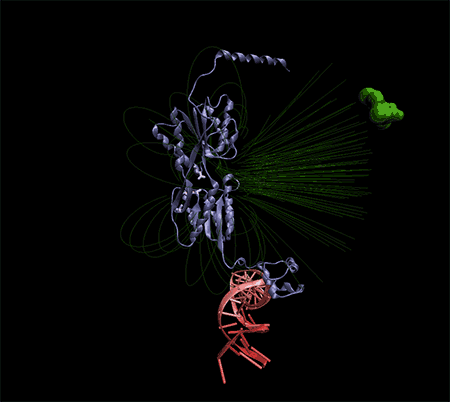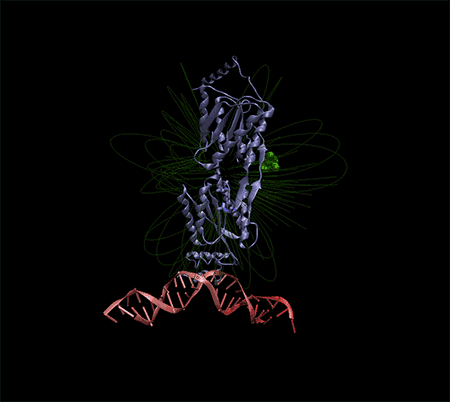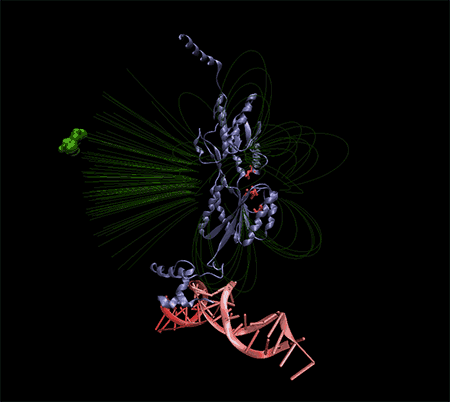
Note: All electrochemical nano biosensors that have a molecular base depend on a very specific system to detect or track their target molecule. The importance of an electrochemical nanobiosensor is to provide a suitable support for connecting the target molecule to the probe and creating an electrical signal that can be measured and read.
In the building of electrochemical biosensors, the minimum parts that are used in a biosensor are: molecular recognition layer and signal transducer which can be connected to a measuring device (device readout) of these signals. DNA is usually a suitable tool as a biosensor because the base pairing reaction between complementary sequences is both specific and stable. In this case, single-stranded probe DNA is immobilized on the detection layer, and then the target DNA reacts with the probe on the surface by pairing. The repetitiveness and unity of DNA structures makes their accumulation on the surface very specific. It is on this surface that the target DNA is taken and the signal is created. Therefore, it is important to immobilize the nucleic acid of the probe while maintaining its initial adhesion strength to detect the target DNA. But how this diagnostic process is measured depends on the method of signal transduction, which may be optical, mechanical, or electrochemical. Optical bisensors that work based on fluorescence light have some characteristics. These types of biosensors are sensitive to molecules per square centimeter. They consist of 7 rows, so that their detection limit is almost made of thousands of probes.
Because the tools in this field (fluorescence biosensor) are complex and expensive . Gene chip technology is more suitable for laboratory work. Gene chips are more suitable in cases where a lot of work needs to be done at the same time, such as transcriptional profiling or single nucleotide polymorphism discovery. But clinical diagnoses usually require the collection of these large amounts of data. They are not. What is important for molecular diagnosis is the reliability of the diagnosis as well as its generality regardless of the order of the game. Therefore, gene chips are not preferred for clinical diagnosis for reasons such as: it is expensive and the device is complicated. Due to other specific reasons , the reading accuracy decreases. Another method to measure the optical signal is the Resonance Plamon Surface method. In this method, a change is made in the refractive index of a thin metal film substrate, which It is due to the absorption of the analyte and it is suitable for target detection in home-home, groove-groove states. In order to be able to detect the target molecule in this method, where a signal is generated, the hybridization signal must be strengthened This action can be enhanced by increasing the amount of material on the surface of the film before or after binding to the target DNA. The Surface Plasmon Resonance (SPR) method is like the expensive and complicated fluorescence method, which is the reason why this method is used more for research work than for routine diagnostic work; One of the optical signal measurement methods that is very specific is the optical reading method, in which single-stranded DNAs are labeled with gold nanoparticles, which easily change color due to hybridization with the sequence of the target game . By using silver staining, DNA analysis can be performed with this optical method in very small plates with high sensitivity, although the use of gold nanoparticles may be expensive, but this method has the necessary sensitivity and simplicity for clinical diagnoses.
One of the methods of reading or measuring the signal is to measure the changes that occur in the immobilized detection layer, which is caused by binding to the target molecule. In this case, Quartz Microbalance Crystal is mostly used. This device is sensitive and can report hybrid production (mating) when it is created. One of the limitations of the QMC method is that this method cannot be performed when the target sample is in the solution phase, but with the advances in this field, i.e. the creation of new methods of sample amplification and amplification, this limitation may be eliminated. Another method of volume measurement is the use of microfabricated cantilevers. In this method, the increase in volume that accompanies hybridization is detected by the deviation of the laser beam from the surface of the cantilever. One of the advantages of this method is that it is suitable for the development of creating linear grooves and that non-specific connections can be corrected with this method. The disadvantages of this method are the expensive and complicated tools and devices used. Electrochemical methods are very suitable for DNA detection because electrochemical reactions directly create an electronic signal and therefore there is no need for expensive converter devices. In addition, since the immobilized game sequence can be limited to only a series of electrode substrates, the act of demonstration (tracking) can be performed by a series of cheap electrochemical analyses . Electrochemical sensors are available to perform clinical or environmental tests on site . The sensitivity of electrochemical signals is based on direct oxidation or catalysis of DNA bases, as well as reduction reactions of reporter molecules or enzymes. Various methods are used to measure the electrochemical signal. The basis of signal measurement in direct electrochemistry of DNA is based on the oxidation and reduction reaction of DNA in a mercury electrode, therefore, the amount of oxidized and reduced DNA is proportional to the amount of DNA that is hybridized with the probe. In addition to the old methods of direct DNA reduction, nowadays a method called stripping adsorption voltammetry of surface nanomolecules is used to select the direct oxidation nanomolecules of DNA, which is a very sensitive method. In the direct electrochemical method, purine bases are oxidized by materials such as carbon, indium oxide (ITO), gold, and polymer-covered electrodes . Although the direct electrochemical method is a very sensitive method, its application is complicated.
Conclusion :
All molecular-based electrochemical nano-biosensors depend on a very specific system to detect or track their target molecule. The importance of an electrochemical nanobiosensor is to provide a suitable support for connecting the target molecule to the probe and creating an electrical signal that can be measured and read.
Researcher and author: Dr. ( Afshin Rashid)
Specialized doctorate in nano-microelectronics