Nanotechnology and modern molecular simulation techniques to a general framework for the interpretation of AFM images, especially the analysis of atomic mechanisms (PhD in Nano-Microelectronics)
Researcher and author: Dr. ( Afshin Rashid)
Note: In nanotechnology, from modern molecular simulation techniques to a general framework for interpreting AFM images, especially the analysis of atomic mechanisms, they create changes in the force measured by a microscope and determine the contrast of the image. produces.
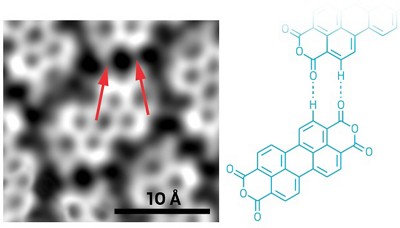
The recent advent of high-resolution imaging and force spectroscopy using atomic force microscopy (AFM) in organic and organic solutions paves the way for imaging a wide range of surfaces and their solvent structure. However, to take full advantage of the high resolution and provide a remarkable new analytical ability, it is important to have a clear understanding of the underlying contrast mechanisms that lead to atomic and molecular separation. Without a theory that connects the measured force to the atomic models of the surface and tip of the microscope, the distillate information from these measurements is limited Molecular dynamics simulations show that the forces acting on the tip of a microscope are the result of direct interaction between a tip and a surface, and are exerted entirely by the structure of water around the tip and surface. The observed force depends on a tip structure, and the balance between potential energy changes is mainly repulsive as the tip approaches the surface and increases entropically, which sterically prevents the occupation of places near the tip and the water surface. Understanding the interaction of these various components, which contribute to the measurement power of the microscope, is critical to interpreting high-resolution images of solution interfaces.
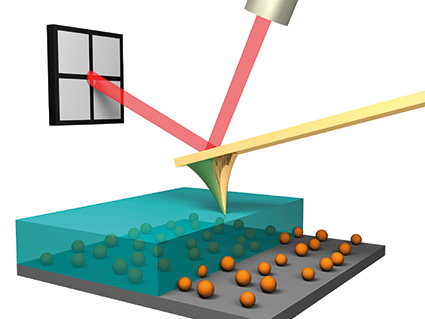
The Atomic Force Microscope (AFM) is one of the best tools for imaging, measuring, and manipulating matter at the nanoscale, and in fact, seeing atoms and molecular bonds up close. This technique produces images that look a bit like a sound player. An atomic-scale needle at the end of the console arm scans the specimen and moves up and down according to the shape and electronic characteristics of the surface. Measuring and recording that deviation produces a three-dimensional representation of the sample molecules.The atomic force microscope uses a sharp probe that moves on the surface of the specimen under study. In the case of the atomic force microscope, there is a tip on the lever that bends between the specimen and the tip. As the cantilever bends, the reflection of the laser light on the optical detector shifts. In this way, the displacement of the Oliver County tip can be measured. Since Kant-Oliver follows Hooke's law in small displacements, the force of all the interaction between the tip and the surface of the specimen can be obtained from Kant-Oliver displacements. And from the force between the atoms of the sample surface and the probe, the distance between the tip and the surface of the sample, or the same height of that part of the sample can be obtained. The movement of the probe on the sample is done by a very precise positioning device.
Conclusion :
In nanotechnology, they develop from modern molecular simulation techniques to a general framework for interpreting AFM images, in particular the analysis of atomic mechanisms that produce force changes measured by a microscope and determine the contrast of an image. Slowly
Researcher and author: Dr. ( Afshin Rashid)
PhD in Nano-Microelectronics